Building a CLSTM cell for 4D Data
LSTM or Long Short-Term Memory is a specialized type of recurrent neural network (RNN) designed to address the vanishing gradient problem often encountered by traditional RNNs when processing long sequences.
Recurrent Neural Networks (RNNs) are a type of neural network designed to work with sequential data. Unlike traditional feedforward networks, RNNs have connections that loop back on themselves, allowing them to maintain a “memory” of past inputs. This memory enables them to process information from previous steps in the sequence, making them well-suited for tasks like natural language processing, speech recognition, and time series analysis. However they are prone to vanishing gradient problem.
LSTMs overcome this by incorporating a memory cell, which allows the network to store and access information over extended periods. Here is a pictoral representation of an LSTM Cell.
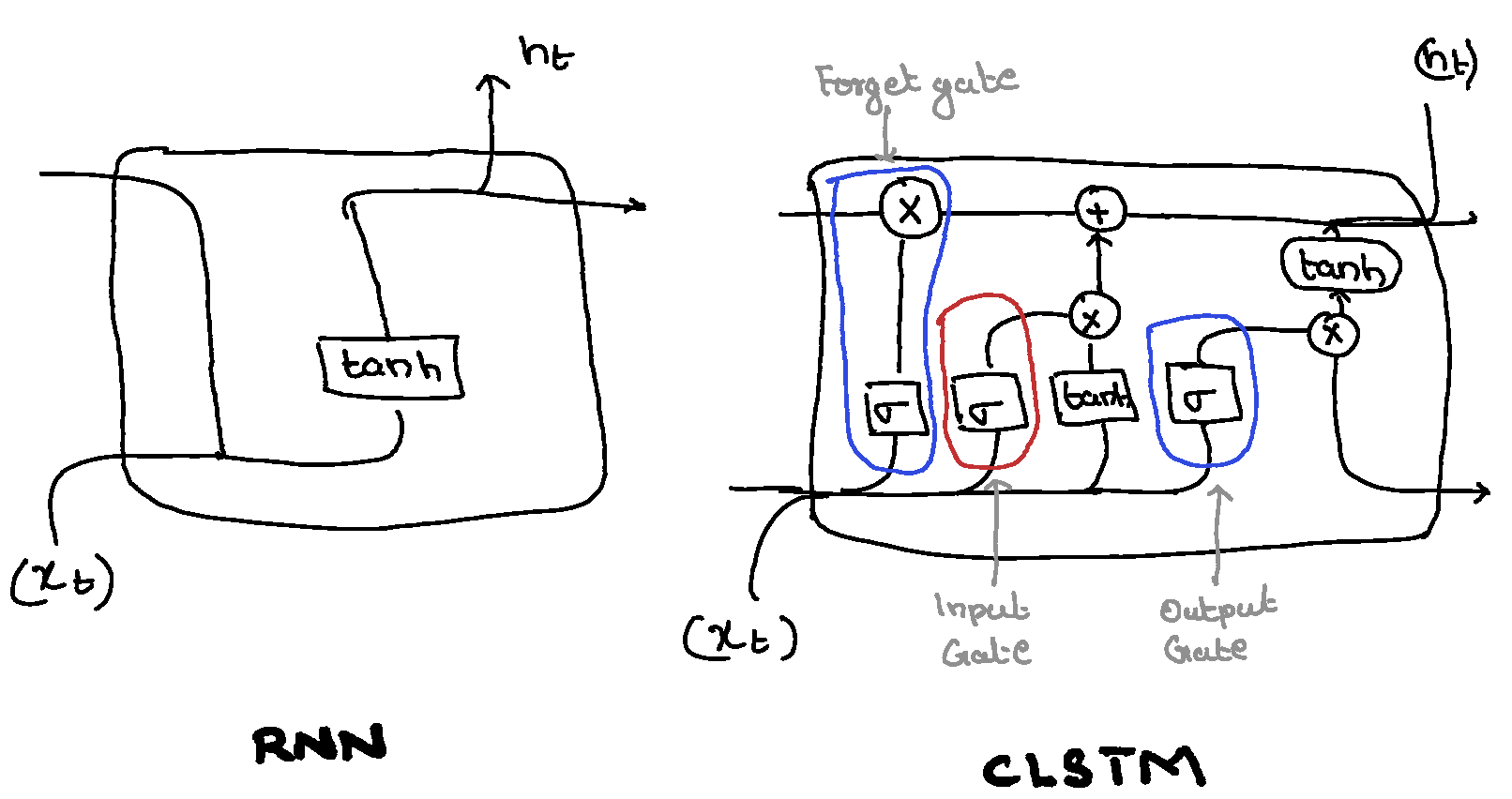
The LSTM cell consists of three gates:
- Input Gate: Determines which new information should be stored in the cell.
- Forget Gate: Decides which information should be discarded from the cell.
- Output Gate: Controls what information from the cell is used to compute the output.
These gates regulate the flow of information into and out of the cell, enabling the network to selectively remember or forget information.
Pytorch implementation
Here is the pytorch implementation of a vanilla LSTM Cell.
import torch
import torch.nn as nn
class LSTMCell(nn.Module):
def __init__(self, input_size, hidden_size):
super(LSTMCell, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
# Forget, Input, and Output gates weights
self.Wf = nn.Linear(input_size + hidden_size, hidden_size)
self.Wi = nn.Linear(input_size + hidden_size, hidden_size)
self.Wc = nn.Linear(input_size + hidden_size, hidden_size)
self.Wo = nn.Linear(input_size + hidden_size, hidden_size)
def forward(self, x, hx):
h_prev, c_prev = hx
# Concatenate input and previous hidden state
combined = torch.cat((x, h_prev), dim=1)
# Forget gate
f_t = torch.sigmoid(self.Wf(combined))
# Input gate
i_t = torch.sigmoid(self.Wi(combined))
# Candidate memory
c_hat_t = torch.tanh(self.Wc(combined))
# Cell state
c_t = f_t * c_prev + i_t * c_hat_t
# Output gate
o_t = torch.sigmoid(self.Wo(combined))
# Hidden state
h_t = o_t * torch.tanh(c_t)
return h_t, c_t
Sample code for running the vanilla LSTM Cell.
input_size = 4
hidden_size = 3
lstm_cell = LSTMCell(input_size, hidden_size)
# Example input (batch_size=1, input_size=4)
x = torch.randn(1, input_size)
# Initial hidden state and cell state
h_prev = torch.zeros(1, hidden_size)
c_prev = torch.zeros(1, hidden_size)
h_t, c_t = lstm_cell(x, (h_prev, c_prev))
print("Next hidden state:", h_t)
print("Next cell state:", c_t)
Converting LSTM into CLSTM
To convert an LSTM cell into a Convolutional LSTM (ConvLSTM), you typically use convolutional layers instead of fully connected layers for processing the input and hidden states. Below is an implementation of a ConvLSTM cell in PyTorch:
import torch
import torch.nn as nn
class ConvLSTMCell(nn.Module):
def __init__(self, input_size, hidden_size, kernel_size, padding=0, bias=True):
super(ConvLSTMCell, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.kernel_size = kernel_size
self.padding = padding
# Define the convolutional layers for the gates
self.Wf = nn.Conv2d(in_channels=input_size + hidden_size,
out_channels=hidden_size,
kernel_size=kernel_size,
padding=padding)
self.Wi = nn.Conv2d(in_channels=input_size + hidden_size,
out_channels=hidden_size,
kernel_size=kernel_size,
padding=padding)
self.Wc = nn.Conv2d(in_channels=input_size + hidden_size,
out_channels=hidden_size,
kernel_size=kernel_size,
padding=padding)
self.Wo = nn.Conv2d(in_channels=input_size + hidden_size,
out_channels=hidden_size,
kernel_size=kernel_size,
padding=padding)
def forward(self, x, hx):
h_prev, c_prev = hx
# Concatenate input and previous hidden state along the channel dimension
combined = torch.cat((x, h_prev), dim=1)
# Forget gate
f_t = torch.sigmoid(self.Wf(combined))
# Input gate
i_t = torch.sigmoid(self.Wi(combined))
# Candidate memory
c_hat_t = torch.tanh(self.Wc(combined))
# Now the operation is a Convolution
c_t = f_t * c_prev + i_t * c_hat_t
# Output gate
o_t = torch.sigmoid(self.Wo(combined))
# Hidden state
h_t = o_t * torch.tanh(c_t)
return h_t, c_t
Sample code to use the CLSTM cell
input_channels = 1
hidden_channels = 3
kernel_size = (3, 3)
conv_lstm_cell = ConvLSTMCell(input_size=input_channels,
hidden_size=hidden_channels,
kernel_size=kernel_size,
padding=1) # Padding to keep the output spatial dimensions same
# Example input (batch_size=1, channels=1, height=4, width=4)
x = torch.randn(1, input_channels, 4, 4)
# Initial hidden state and cell state
h_prev = torch.zeros(1, hidden_channels, 4, 4)
c_prev = torch.zeros(1, hidden_channels, 4, 4)
h_t, c_t = conv_lstm_cell(x, (h_prev, c_prev))
print("Next hidden state shape:", h_t.shape)
print("Next cell state shape:", c_t.shape)
Preparing CLSTM for Handling 3D Data
When dealing with 3D data over time, the input shape follows the format:
- b: batch size
- t: number of time frames
- c: number of channels
- d: depth
- h: height
- w: width
To effectively process this type of data, the clstm3D class will execute the cell for t time steps in the forward function.
class CLSTMCell3D(nn.Module):
def __init__(self, input_channels, hidden_channels, kernel_size):
super(CLSTMCell3D, self).__init__()
self.input_channels = input_channels
self.hidden_channels = hidden_channels
self.kernel_size = kernel_size
self.padding = kernel_size // 2 # to preserve spatial dimensions
self.conv = nn.Conv3d(
in_channels=input_channels + hidden_channels,
out_channels=4 * hidden_channels,
kernel_size=kernel_size,
padding=self.padding
)
def forward(self, x, h, c):
# Concatenate input and hidden state along the channel dimension
combined = torch.cat([x, h], dim=1)
conv_output = self.conv(combined)
i, f, o, g = torch.chunk(conv_output, 4, dim=1)
i = torch.sigmoid(i) # input gate
f = torch.sigmoid(f) # forget gate
o = torch.sigmoid(o) # output gate
g = torch.tanh(g) # cell gate
c_next = f * c + i * g
h_next = o * torch.tanh(c_next)
return h_next, c_next
class CLSTM3D(nn.Module):
"""ConvLSTM module for 3D data."""
def __init__(self, input_dim, hidden_dim, kernel_size, num_layers, bias=True, batch_first=True):
super(CLSTM3D, self).__init__()
self.num_layers = num_layers
self.hidden_dim = hidden_dim
self.batch_first = batch_first
self.lstm_cells = nn.ModuleList([
CLSTMCell3D(input_dim if i == 0 else hidden_dim, hidden_dim, kernel_size)
for i in range(num_layers)
])
def forward(self, x):
# Initialize hidden and cell states
b, t, c, d, h, w = x.size()
hidden_states = [None] * self.num_layers
outputs = []
for t_step in range(t):
x_t = x[:, t_step] if self.batch_first else x[t_step]
for layer_idx, lstm_cell in enumerate(self.lstm_cells):
if hidden_states[layer_idx] is None:
hidden_states[layer_idx] = (torch.zeros(b, self.hidden_dim, d, h, w).to(x.device),
torch.zeros(b, self.hidden_dim, d, h, w).to(x.device))
hidden_state, cell_state = lstm_cell(x_t, hidden_states[layer_idx][0], hidden_states[layer_idx][1])
hidden_states[layer_idx] = (hidden_state, cell_state)
x_t = hidden_state
outputs.append(x_t.unsqueeze(1))
return torch.cat(outputs, dim=1)
Implementation of CLSTM in UNet3D
For more details on the implementation of CLSTM in 3D, you can check my project posting 4D Time-Resolved UNet.
The code for the CLSTM cell is open-sourced and can be found on GitHub.
You can also explore the full implementation of the 4D time-resolved UNet in this GitHub repository.
Limitations
One of the limitations of this approach is the larger memory footprint. LSTM cells are designed to store and manage higher-dimensional data, especially when dealing with 3D data over time. The complete implementation of the 4D time-resolved UNet was able to run on an A30 GPU with 40 GB VRAM, albeit with a lower patch size.
Enjoy Reading This Article?
Here are some more articles you might like to read next: